Change breaks in a "classIntervals" object
getBclustClassIntervals.RdBecause "classIntervals" objects of style "hclust" or "bclust" contain hierarchical classification trees in their "par" attribute, different numbers of classes can be chosen without repeating the initial classification. This function accesses the "par" attribute and modifies the "brks" member of the returned "classIntervals" object.
Examples
if (!require("spData", quietly=TRUE)) {
message("spData package needed for examples")
run <- FALSE
} else {
run <- TRUE
}
if (run) {
data(jenks71, package="spData")
pal1 <- c("wheat1", "red3")
opar <- par(mfrow=c(2,2))
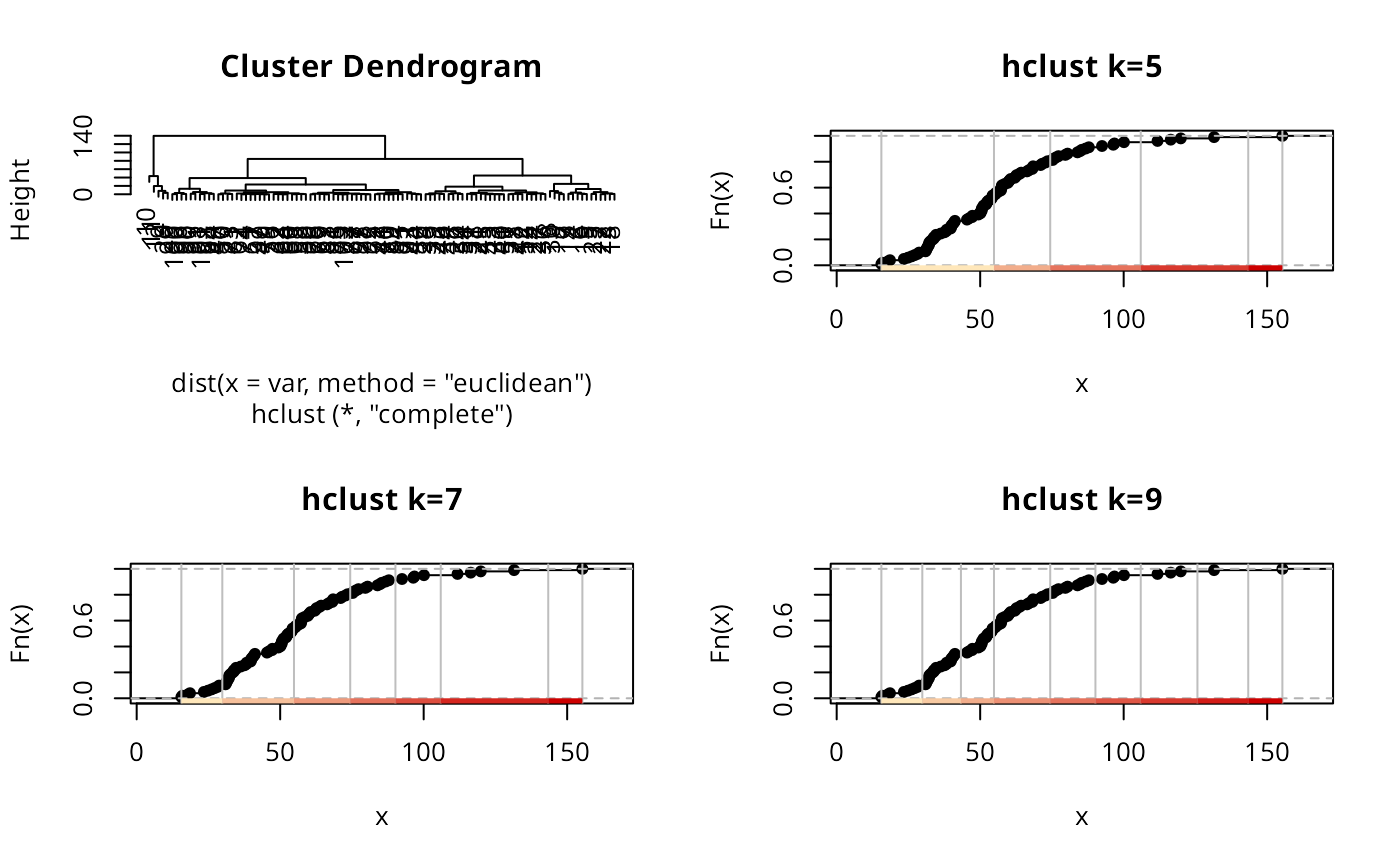
hCI5 <- classIntervals(jenks71$jenks71, n=5, style="hclust", method="complete")
plot(attr(hCI5, "par"))
plot(hCI5, pal=pal1, main="hclust k=5")
plot(getHclustClassIntervals(hCI5, k=7), pal=pal1, main="hclust k=7")
plot(getHclustClassIntervals(hCI5, k=9), pal=pal1, main="hclust k=9")
par(opar)
}
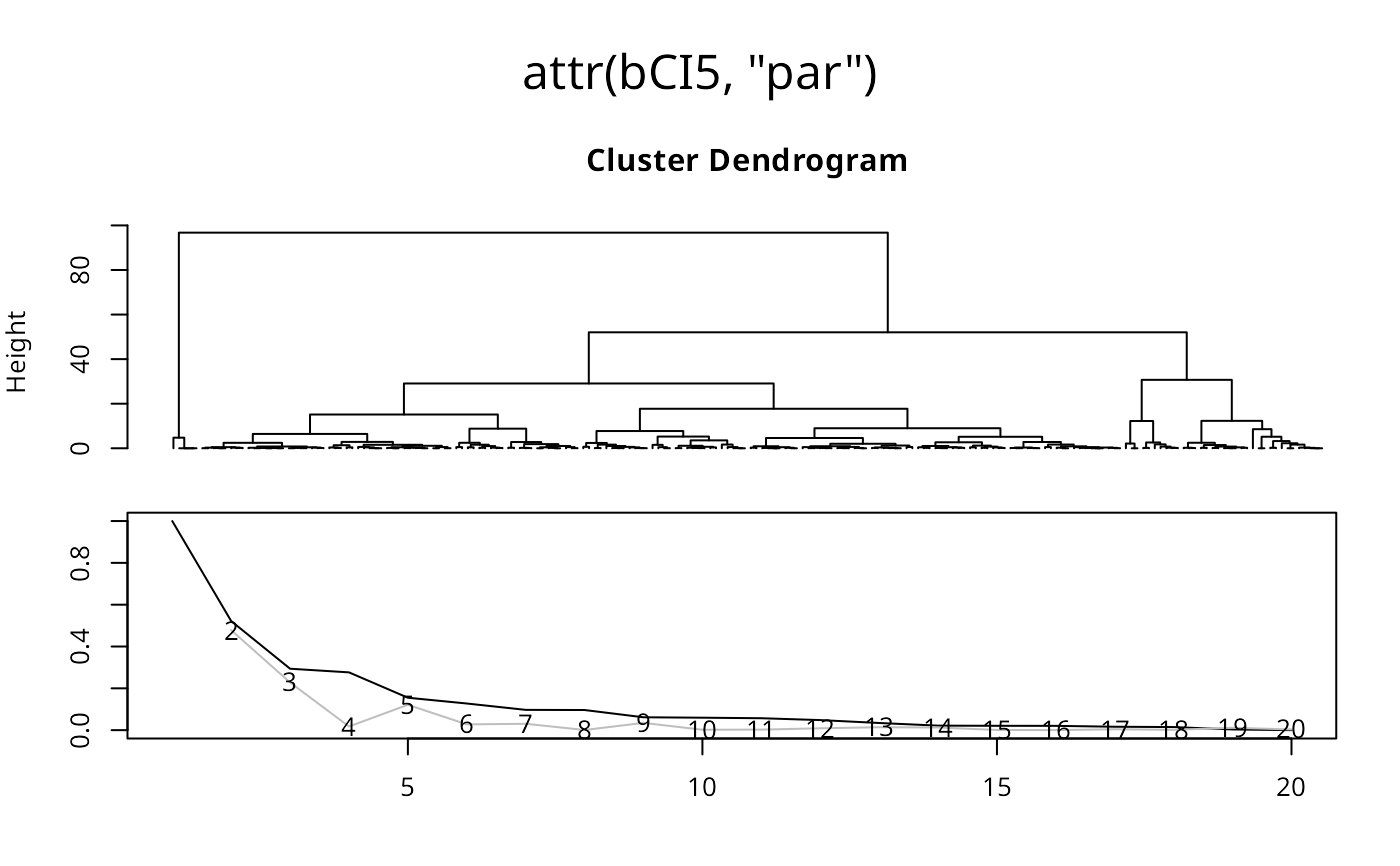
 if (run) {
set.seed(1)
bCI5 <- classIntervals(jenks71$jenks71, n=5, style="bclust")
plot(attr(bCI5, "par"))
}
#> Committee Member: 1(1) 2(1) 3(1) 4(1) 5(1) 6(1) 7(1) 8(1) 9(1) 10(1)
#> Computing Hierarchical Clustering
if (run) {
set.seed(1)
bCI5 <- classIntervals(jenks71$jenks71, n=5, style="bclust")
plot(attr(bCI5, "par"))
}
#> Committee Member: 1(1) 2(1) 3(1) 4(1) 5(1) 6(1) 7(1) 8(1) 9(1) 10(1)
#> Computing Hierarchical Clustering
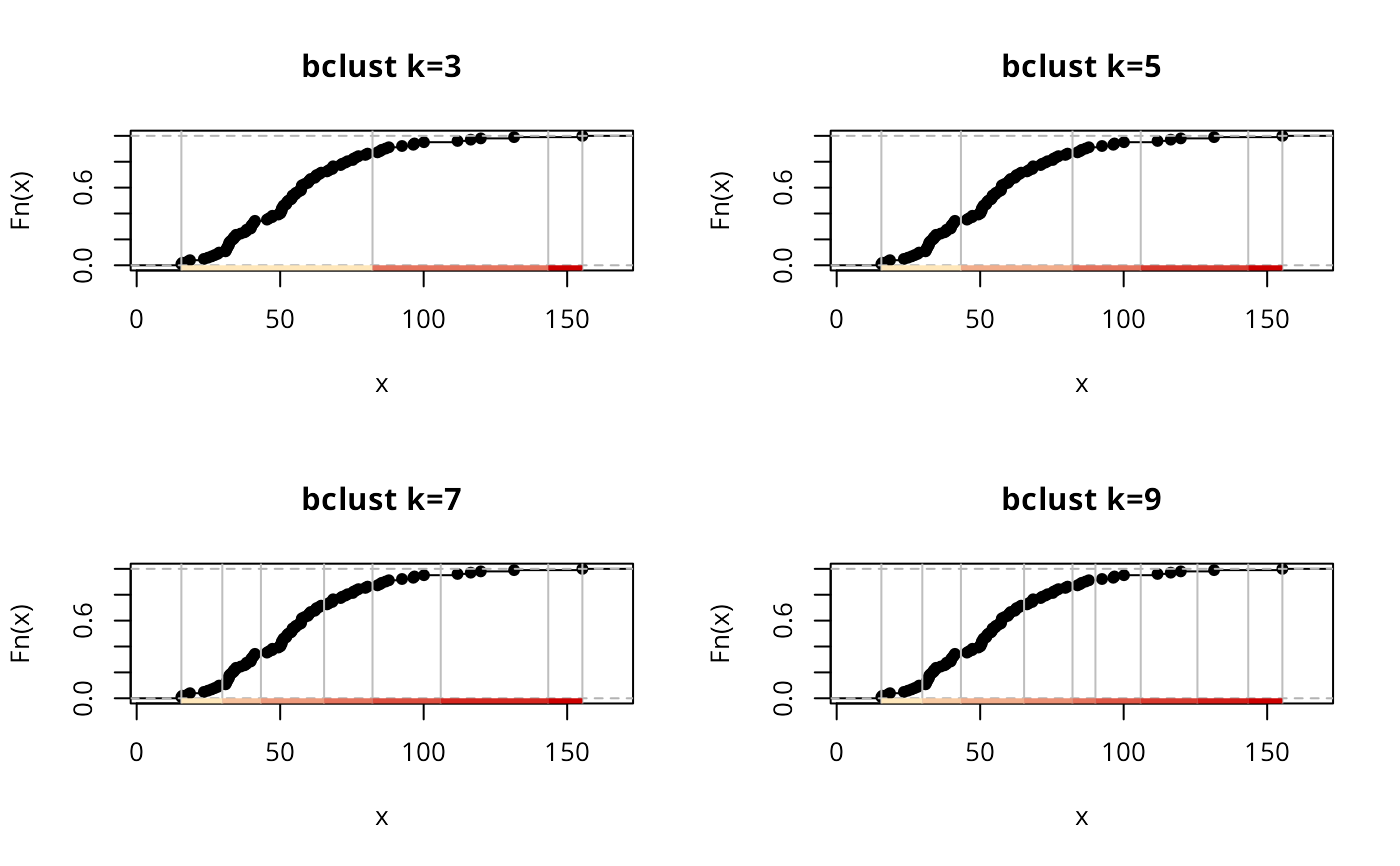
 if (run) {
opar <- par(mfrow=c(2,2))
plot(getBclustClassIntervals(bCI5, k=3), pal=pal1, main="bclust k=3")
plot(bCI5, pal=pal1, main="bclust k=5")
plot(getBclustClassIntervals(bCI5, k=7), pal=pal1, main="bclust k=7")
plot(getBclustClassIntervals(bCI5, k=9), pal=pal1, main="bclust k=9")
par(opar)
}
if (run) {
opar <- par(mfrow=c(2,2))
plot(getBclustClassIntervals(bCI5, k=3), pal=pal1, main="bclust k=3")
plot(bCI5, pal=pal1, main="bclust k=5")
plot(getBclustClassIntervals(bCI5, k=7), pal=pal1, main="bclust k=7")
plot(getBclustClassIntervals(bCI5, k=9), pal=pal1, main="bclust k=9")
par(opar)
}