“The Problem of Spatial Autocorrelation:” forty years on
Roger Bivand
Source:vignettes/CO69.Rmd
CO69.RmdIntroduction
Cliff and Ord (1969), published forty years ago, marked a turning point in the treatment of spatial autocorrelation in quantitative geography. It provided the framework needed by any applied researcher to attempt an implementation for a different system, possibly using a different programming language. In this spirit, here we examine how spatial weights have been represented in implementations and may be reproduced, how the tabulated results in the paper may be reproduced, and how they may be extended to cover simulation.
One of the major assertions of Cliff and Ord (1969) is that their statistic advances the measurement of spatial autocorrelation with respect to Moran (1950) and Geary (1954) because a more general specification of spatial weights could be used. This more general form has implications both for the preparation of the weights themselves, and for the calculation of the measures. We will look at spatial weights first, before moving on to consider the measures presented in the paper and some of their subsequent developments. Before doing this, we will put together a data set matching that used in Cliff and Ord (1969). They provide tabulated data for the counties of the Irish Republic, but omit Dublin from analyses. A shapefile included in this package, kindly made available by Michael Tiefelsdorf, is used as a starting point:
library(spdep)
eire <- as(sf::st_read(system.file("shapes/eire.gpkg", package="spData")[1]), "Spatial")
row.names(eire) <- as.character(eire$names)
#proj4string(eire) <- CRS("+proj=utm +zone=30 +ellps=airy +units=km")
class(eire)## [1] "SpatialPolygonsDataFrame"
## attr(,"package")
## [1] "sp"
names(eire)## [1] "A" "towns" "pale" "size" "ROADACC" "OWNCONS" "POPCHG"
## [8] "RETSALE" "INCOME" "names"and read into a SpatialPolygonsDataFrame — classes used for handling spatial data in are fully described in Roger S. Bivand, Pebesma, and Gómez-Rubio (2008). To this we need to add the data tabulated in the paper in Table 2,1 p. 40, here in the form of a text file with added rainfall values from Table 9, p. 49:
fn <- system.file("etc/misc/geary_eire.txt", package="spdep")[1]
ge <- read.table(fn, header=TRUE)
names(ge)## [1] "serlet" "county" "pagval2_10" "pagval10_50"
## [5] "pagval50p" "cowspacre" "ocattlepacre" "pigspacre"
## [9] "sheeppacre" "townvillp" "carspcap" "radiopcap"
## [13] "retailpcap" "psinglem30_34" "rainfall"Since we assigned the county names as feature identifiers when reading the shapefiles, we do the same with the extra data, and combine the objects:
row.names(ge) <- as.character(ge$county)
all.equal(row.names(ge), row.names(eire))## [1] TRUE
eire_ge <- cbind(eire, ge)Finally, we need to drop the Dublin county omitted in the analyses conducted in Cliff and Ord (1969):
## [1] 25To double-check our data, let us calculate the sample Beta coefficients, using the formulae given in the paper for sample moments:
skewness <- function(z) {z <- scale(z, scale=FALSE); ((sum(z^3)/length(z))^2)/((sum(z^2)/length(z))^3)}
kurtosis <- function(z) {z <- scale(z, scale=FALSE); (sum(z^4)/length(z))/((sum(z^2)/length(z))^2)}These differ somewhat from the ways in which skewness and kurtosis are computed in modern statistical software, see for example Joanes and Gill (1998). However, for our purposes, they let us reproduce Table 3, p. 42:
## pagval2_10 pagval10_50 pagval50p cowspacre ocattlepacre
## 1.675429 1.294978 0.000382 1.682094 0.086267
## pigspacre sheeppacre townvillp carspcap radiopcap
## 1.138387 1.842362 0.472748 0.011111 0.342805
## retailpcap psinglem30_34
## 0.002765 0.068169## pagval2_10 pagval10_50 pagval50p cowspacre ocattlepacre
## 3.790 4.331 1.508 4.294 2.985
## pigspacre sheeppacre townvillp carspcap radiopcap
## 3.754 4.527 2.619 1.865 2.730
## retailpcap psinglem30_34
## 2.188 2.034## pagval2_10 cowspacre pigspacre sheeppacre
## 0.68801 0.17875 0.00767 0.04184## pagval2_10 cowspacre pigspacre sheeppacre
## 2.883 2.799 2.212 2.421Using the tabulated value of \(23.6\) for the percentage of agricultural
holdings above 50 in 1950 in Waterford, the skewness and kurtosis cannot
be reproduced, but by comparison with the
irishdata dataset in , it turns out that the
value should rather be \(26.6\), which
yields the tabulated skewness and kurtosis values.
Before going on, the variables considered are presented in Table \[vars\].
| variable | description |
|---|---|
| pagval2_10 | Percentage number agricultural holdings in valuation group £2–£10 (1950) |
| pagval10_50 | Percentage number agricultural holdings in valuation group £10–£50 (1950) |
| pagval50p | Percentage number agricultural holdings in valuation group above £50 (1950) |
| cowspacre | Milch cows per 1000 acres crops and pasture (1952) |
| ocattlepacre | Other cattle per 1000 acres crops and pasture (1952) |
| pigspacre | Pigs per 1000 acres crops and pasture (1952) |
| sheeppacre | Sheep per 1000 acres crops and pasture (1952) |
| townvillp | Town and village population as percentage of total (1951) |
| carspcap | Private cars registered per 1000 population (1952) |
| radiopcap | Radio licences per 1000 population (1952) |
| retailpcap | Retail sales £ per person (1951) |
| psinglem30_34 | Single males as percentage of all males aged 30–34 (1951) |
| rainfall | Average of rainfall for stations in Ireland, 1916–1950, mm |
Spatial weights
As a basis for comparison, we will first read the unstandardised weighting matrix given in Table A1, p. 54, of the paper, reading a file corrected for the misprint giving O rather than D as a neighbour of V:
fn <- system.file("etc/misc/unstand_sn.txt", package="spdep")[1]
unstand <- read.table(fn, header=TRUE)
summary(unstand)## from to weight
## Length:110 Length:110 Min. :0.000600
## Class :character Class :character 1st Qu.:0.003225
## Mode :character Mode :character Median :0.007550
## Mean :0.007705
## 3rd Qu.:0.010225
## Max. :0.032400In the file, the counties are represented by their serial letters, so ordering and conversion to integer index representation is required to reach a representation similar to that of the SpatialStats module for spatial neighbours:
class(unstand) <- c("spatial.neighbour", class(unstand))
of <- ordered(unstand$from)
attr(unstand, "region.id") <- levels(of)
unstand$from <- as.integer(of)
unstand$to <- as.integer(ordered(unstand$to))
attr(unstand, "n") <- length(unique(unstand$from))Having done this, we can change its representation to a
listw object, assigning an appropriate style
(generalised binary) for unstandardised values:
lw_unstand <- sn2listw(unstand)
lw_unstand$style <- "B"
lw_unstand## Characteristics of weights list object:
## Neighbour list object:
## Number of regions: 25
## Number of nonzero links: 110
## Percentage nonzero weights: 17.6
## Average number of links: 4.4
##
## Weights style: B
## Weights constants summary:
## n nn S0 S1 S2
## B 25 625 0.8476 0.01871808 0.1229232Note that the values of S0, S1, and S2 correspond closely with those given on page 42 of the paper, \(0.84688672\), \(0.01869986\) and \(0.12267319\). The discrepancies appear to be due to rounding in the printed table of weights.
The contiguous neighbours represented in this object ought to match
those found using poly2nb. However, we see
that the reproduced contiguities have a smaller link count:
nb <- poly2nb(eire_ge1)
nb## Neighbour list object:
## Number of regions: 25
## Number of nonzero links: 108
## Percentage nonzero weights: 17.28
## Average number of links: 4.32The missing link is between Clare and Kerry, perhaps by the Tarbert–Killimer ferry, but the counties are not contiguous, as Figure \[plot\_nb\] shows:
xx <- diffnb(nb, lw_unstand$neighbours, legacy=TRUE, verbose=TRUE)## Neighbour difference for region id: Clare in relation to id: Kerry
## Neighbour difference for region id: Kerry in relation to id: Clare
plot(eire_ge1, border="grey60")
plot(nb, coordinates(eire_ge1), add=TRUE, pch=".", lwd=2)
plot(xx, coordinates(eire_ge1), add=TRUE, pch=".", lwd=2, col=3)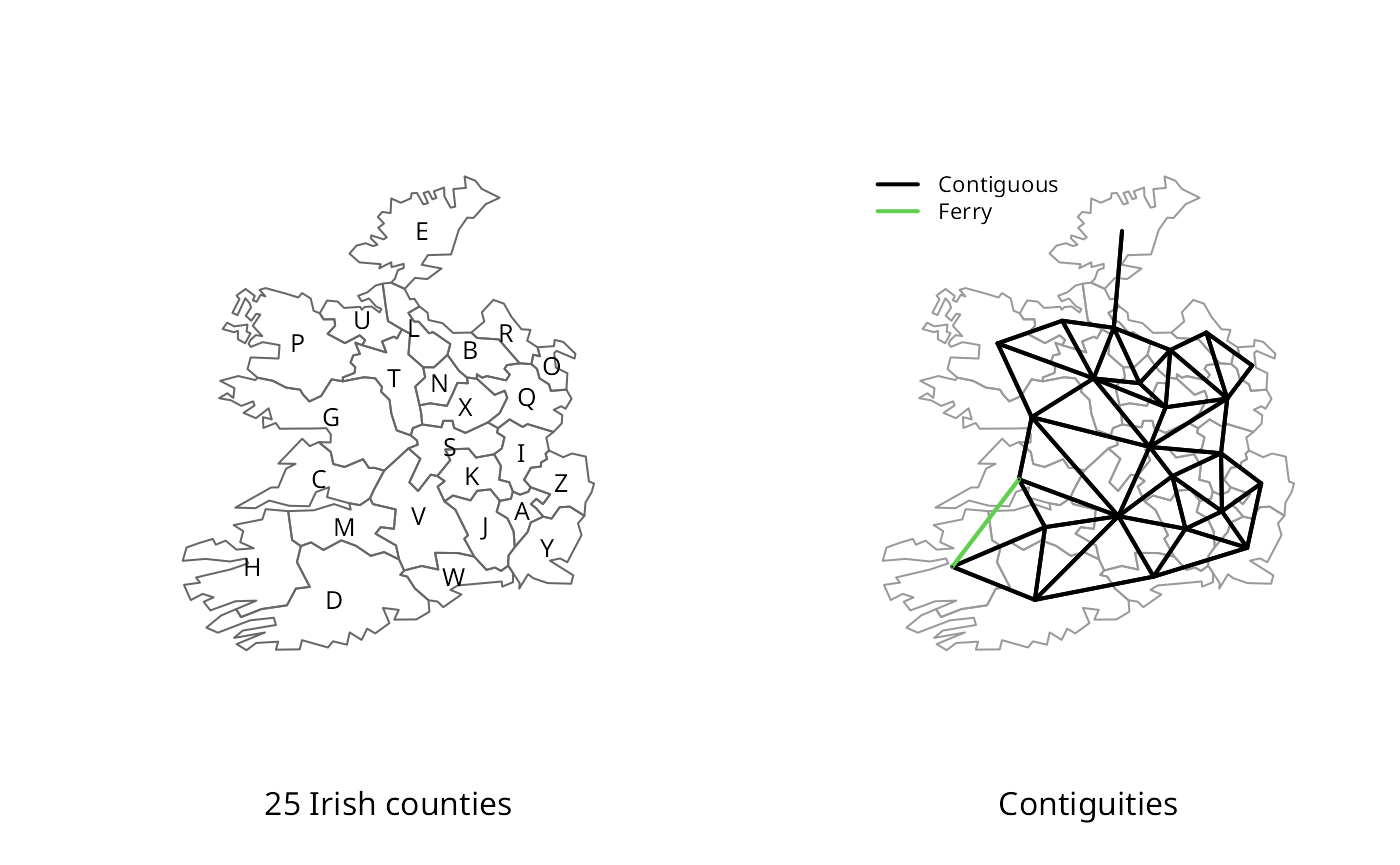
County boundaries and contiguities
An attempt has also been made to reproduce the generalised weights
for 25 Irish counties reported by Cliff and Ord
(1969), after Dublin is omitted. Reproducing the inverse distance
component \(d_{ij}^{-1}\) of the
generalised weights \(d_{ij}^{-1}
\beta_{i(j)}\) is eased by the statement in Cliff and Ord (1973, 55) that the points chosen
to represent the counties were their “geographic centres,” so not very
different from the centroids yielded by applying a chosen computational
geometry function. The distance metric is not given, and may have been
in kilometers or miles — both were tried, but the results were not
sensitive to the difference as it applies equally across the weights;
miles are used here. Computing the proportion of shared distance measure
\(\beta_{i(j)}\) is harder, because it
requires the availability of the full topology of the input polygons.
Roger S. Bivand, Pebesma, and Gómez-Rubio (2008,
244) show how to employ the vect2neigh
function (written by Markus Neteler) in the package when using GRASS GIS
vector handling to create a full topology from spaghetti vector data and
to extract border segment lengths; a similar approach also is mentioned
there using ArcGIS coverages for the same purpose. GRASS was used to
create the topology, and next the proportion of shared distance measure
was calculated.
library(terra)
v_eire_ge1 <-vect(eire_ge1)
SG <- rasterize(v_eire_ge1, rast(nrows=70, ncols=50, extent=ext(v_eire_ge1)), field="county")
library(rgrass)
grass_home <- "/home/rsb/topics/grass/g820/grass82"
initGRASS(grass_home, home=tempdir(), SG=SG, override=TRUE)
write_VECT(v_eire_ge1, "eire", flags=c("o", "overwrite"))
res <- vect2neigh("eire", ID="serlet")
res$length <- res$length*1000
attr(res, "external") <- attr(res, "external")*1000
attr(res, "total") <- attr(res, "total")*1000
grass_borders <- sn2listw(res)
raw_borders <- grass_borders$weights
int_tot <- attr(res, "total") - attr(res, "external")
prop_borders <- lapply(1:length(int_tot), function(i) raw_borders[[i]]/int_tot[i])
dlist <- nbdists(grass_borders$neighbours, coordinates(eire_ge1))
inv_dlist <- lapply(dlist, function(x) 1/(x/1.609344))
combo_km <- lapply(1:length(inv_dlist), function(i) inv_dlist[[i]]*prop_borders[[i]])
combo_km_lw <- nb2listw(grass_borders$neighbours, glist=combo_km, style="B")
summary(combo_km_lw)## Characteristics of weights list object:
## Neighbour list object:
## Number of regions: 25
## Number of nonzero links: 108
## Percentage nonzero weights: 17.28
## Average number of links: 4.32
## Link number distribution:
##
## 1 2 3 4 5 6 7 8
## 1 2 5 5 8 1 2 1
## 1 least connected region:
## E with 1 link
## 1 most connected region:
## V with 8 links
##
## Weights style: B
## Weights constants summary:
## n nn S0 S1.5 S2
## B 25 625 0.9083144 0.02191845 0.1427746To compare, we need to remove the Tarbert–Killimer ferry link manually, and view the summary of the original weights, as well as a correlation coefficient between these and the reconstructed weights. Naturally, unless the boundary coordinates used here are identical with those in the original analysis, presumably measured by hand, small differences will occur.
red_lw_unstand <- lw_unstand
Clare <- which(attr(lw_unstand, "region.id") == "C")
Kerry <- which(attr(lw_unstand, "region.id") == "H")
Kerry_in_Clare <- which(lw_unstand$neighbours[[Clare]] == Kerry)
Clare_in_Kerry <- which(lw_unstand$neighbours[[Kerry]] == Clare)
red_lw_unstand$neighbours[[Clare]] <- red_lw_unstand$neighbours[[Clare]][-Kerry_in_Clare]
red_lw_unstand$neighbours[[Kerry]] <- red_lw_unstand$neighbours[[Kerry]][-Clare_in_Kerry]
red_lw_unstand$weights[[Clare]] <- red_lw_unstand$weights[[Clare]][-Kerry_in_Clare]
red_lw_unstand$weights[[Kerry]] <- red_lw_unstand$weights[[Kerry]][-Clare_in_Kerry]
summary(red_lw_unstand)## Characteristics of weights list object:
## Neighbour list object:
## Number of regions: 25
## Number of nonzero links: 108
## Percentage nonzero weights: 17.28
## Average number of links: 4.32
## Link number distribution:
##
## 1 2 3 4 5 6 7 8
## 1 2 5 5 8 1 2 1
## 1 least connected region:
## E with 1 link
## 1 most connected region:
## V with 8 links
##
## Weights style: B
## Weights constants summary:
## n nn S0 S1 S2
## B 25 625 0.8437 0.01870287 0.1222501## [1] 0.969543Even though the differences in the general weights, for identical contiguities, are so small, the consequences for the measure of spatial autocorrelation are substantial, Here we use the fifth variable, other cattle per 1000 acres crops and pasture (1952), and see that the reconstructed weights seem to “reveal” more autocorrelation than the original weights.
flatten <- function(x, digits=3, statistic="I") {
res <- c(format(x$estimate, digits=digits),
format(x$statistic, digits=digits),
format.pval(x$p.value, digits=digits))
res <- matrix(res, ncol=length(res))
colnames(res) <- paste(c("", "E", "V", "SD_", "P_"), "I", sep="")
rownames(res) <- deparse(substitute(x))
res
}
`reconstructed weights` <- moran.test(eire_ge1$ocattlepacre, combo_km_lw)
`original weights` <- moran.test(eire_ge1$ocattlepacre, red_lw_unstand)
print(rbind(flatten(`reconstructed weights`), flatten(`original weights`)), quote=FALSE)## I EI VI SD_I P_I
## reconstructed weights 0.3203 -0.0417 0.0225 2.41 0.00792
## original weights 0.2779 -0.0417 0.0223 2.14 0.0161Measures of spatial autocorrelation
Our targets for reproduction are Tables 4 and 5 in Cliff and Ord (1969, 43–44), the first containing standard deviates under normality and randomisation for the original Moran measure with binary weights, the original Geary measure with binary weights, the proposed measure with unstandardised general weights, and the proposed measure with row-standardised general weights. In addition, four variables were log-transformed on the basis of the skewness and kurtosis results presented above. We carry out the transformation of these variables, and generate additional binary and row-standardised general spatial weights objects — note that the weights constants for the row-standardised general weights agree with those given on p. 42 in the paper, after allowing for small differences due to rounding in the weights values displayed in the paper (p. 54):
eire_ge1$ln_pagval2_10 <- log(eire_ge1$pagval2_10)
eire_ge1$ln_cowspacre <- log(eire_ge1$cowspacre)
eire_ge1$ln_pigspacre <- log(eire_ge1$pigspacre)
eire_ge1$ln_sheeppacre <- log(eire_ge1$sheeppacre)
vars <- c("pagval2_10", "ln_pagval2_10", "pagval10_50", "pagval50p",
"cowspacre", "ln_cowspacre", "ocattlepacre", "pigspacre",
"ln_pigspacre", "sheeppacre", "ln_sheeppacre", "townvillp",
"carspcap", "radiopcap", "retailpcap", "psinglem30_34")
nb_B <- nb2listw(lw_unstand$neighbours, style="B")
nb_B## Characteristics of weights list object:
## Neighbour list object:
## Number of regions: 25
## Number of nonzero links: 110
## Percentage nonzero weights: 17.6
## Average number of links: 4.4
##
## Weights style: B
## Weights constants summary:
## n nn S0 S1 S2
## B 25 625 110 220 2176
lw_std <- nb2listw(lw_unstand$neighbours, glist=lw_unstand$weights, style="W")
lw_std## Characteristics of weights list object:
## Neighbour list object:
## Number of regions: 25
## Number of nonzero links: 110
## Percentage nonzero weights: 17.6
## Average number of links: 4.4
##
## Weights style: W
## Weights constants summary:
## n nn S0 S1 S2
## W 25 625 25 15.84089 103.6197The standard representation of the measures is:
\[I = \frac{n}{\sum_{i=1}^{n}\sum_{j=1}^{n}w_{ij}} \frac{\sum_{i=1}^{n}\sum_{j=1}^{n}w_{ij}(x_i-\bar{x})(x_j-\bar{x})}{\sum_{i=1}^{n}(x_i - \bar{x})^2}\]
for Moran’s \(I\) — in the paper termed the proposed statistic, and for Geary’s \(C\):
\[C = \frac{(n-1)}{2\sum_{i=1}^{n}\sum_{j=1}^{n}w_{ij}} \frac{\sum_{i=1}^{n}\sum_{j=1}^{n}w_{ij}(x_i-x_j)^2}{\sum_{i=1}^{n}(x_i - \bar{x})^2}\]
where \(x_i, i=1, \ldots, n\) are
\(n\) observations on the numeric
variable of interest, and \(w_{ij}\)
are the spatial weights. In order to reproduce the standard deviates
given in the paper, it is sufficient to apply
moran.test to the variables with three
different spatial weights objects, and two different values of the
randomisation= argument. In addition,
geary.test is applied to a single spatial
weights objects, and two different values of the
randomisation= argument.
system.time({
MoranN <- lapply(vars, function(x) moran.test(eire_ge1[[x]], listw=nb_B, randomisation=FALSE))
MoranR <- lapply(vars, function(x) moran.test(eire_ge1[[x]], listw=nb_B, randomisation=TRUE))
GearyN <- lapply(vars, function(x) geary.test(eire_ge1[[x]], listw=nb_B, randomisation=FALSE))
GearyR <- lapply(vars, function(x) geary.test(eire_ge1[[x]], listw=nb_B, randomisation=TRUE))
Prop_unstdN <- lapply(vars, function(x) moran.test(eire_ge1[[x]], listw=lw_unstand, randomisation=FALSE))
Prop_unstdR <- lapply(vars, function(x) moran.test(eire_ge1[[x]], listw=lw_unstand, randomisation=TRUE))
Prop_stdN <- lapply(vars, function(x) moran.test(eire_ge1[[x]], listw=lw_std, randomisation=FALSE))
Prop_stdR <- lapply(vars, function(x) moran.test(eire_ge1[[x]], listw=lw_std, randomisation=TRUE))
})## user system elapsed
## 0.12 0.00 0.12
res <- sapply(c("MoranN", "MoranR", "GearyN", "GearyR", "Prop_unstdN", "Prop_unstdR", "Prop_stdN", "Prop_stdR"), function(x) sapply(get(x), "[[", "statistic"))
rownames(res) <- vars
ores <- res[,c(1,2,5:8)]In order to conduct 8 different tests on 16 variables, we use
lapply on the list of variables in the
specified order, then sapply on a list of
output objects by name to generate a table in the same row and column
order as the original (we save a copy of six columns for comparison with
bootstrap results below):
## MoranN MoranR GearyN GearyR Prop_unstdN Prop_unstdR Prop_stdN Prop_stdR
## pagval2_10 3.7851 3.8779 4.3142 3.9016 3.3307 3.4159 3.9276 4.0292
## ln_pagval2_10 4.0965 4.1074 4.0841 4.0343 3.5795 3.5894 4.1278 4.1393
## pagval10_50 1.0899 1.1316 2.7511 2.3760 1.3348 1.3882 1.5127 1.5738
## pagval50p 5.2011 5.0555 4.0178 4.7194 4.6604 4.5247 4.8823 4.7387
## cowspacre 5.1969 5.3907 4.3531 3.7709 4.1379 4.2991 4.7274 4.9135
## ln_cowspacre 5.2420 5.2455 3.9211 3.9085 4.2007 4.2037 4.6532 4.6566
## ocattlepacre 0.5565 0.5593 -0.1707 -0.1668 2.1366 2.1478 1.9219 1.9320
## pigspacre 2.4807 2.5393 2.2928 2.0802 2.8312 2.9010 3.1908 3.2703
## ln_pigspacre 2.3015 2.2724 2.0520 2.1893 2.5171 2.4839 2.8460 2.8081
## sheeppacre 1.0188 1.0630 0.8689 0.7387 1.7398 1.8187 1.4792 1.5470
## ln_sheeppacre 1.2930 1.2827 1.5156 1.5767 2.3708 2.3511 2.0374 2.0203
## townvillp 2.2759 2.2681 2.5475 2.5902 1.2148 1.2104 1.6275 1.6216
## carspcap 4.4927 4.4015 3.2247 3.5992 3.8897 3.8075 4.1826 4.0934
## radiopcap 0.3156 0.3153 1.2294 1.2348 0.5915 0.5909 0.7857 0.7849
## retailpcap 3.4985 3.4524 3.1303 3.3497 2.9291 2.8888 3.0346 2.9926
## psinglem30_34 2.7349 2.6895 2.3382 2.5519 2.7541 2.7065 2.7078 2.6605The values of the standard deviates agree with those in Table 4 in the original paper, with the exception of those for the proposed statistic with standardised weights under normality for all untransformed variables. We can see what has happened by substituting the weights constants for the standardised weights with those for unstandardised weights:
wc_unstd <- spweights.constants(lw_unstand)
wrong_N_sqVI <- sqrt((wc_unstd$nn*wc_unstd$S1 - wc_unstd$n*wc_unstd$S2 + 3*wc_unstd$S0*wc_unstd$S0)/((wc_unstd$nn-1)*wc_unstd$S0*wc_unstd$S0)-((-1/(wc_unstd$n-1))^2))
raw_data <- grep("^ln_", vars, invert=TRUE)
I <- sapply(Prop_stdN, function(x) x$estimate[1])[raw_data]
EI <- sapply(Prop_stdN, function(x) x$estimate[2])[raw_data]
res <- (I - EI)/wrong_N_sqVI
names(res) <- vars[raw_data]
print(formatC(res, format="f", digits=4), quote=FALSE)## pagval2_10 pagval10_50 pagval50p cowspacre ocattlepacre pigspacre
## 3.8836 1.4957 4.8276 4.6744 1.9003 3.1550
## sheeppacre townvillp carspcap radiopcap retailpcap psinglem30_34
## 1.4627 1.6093 4.1357 0.7769 3.0006 2.6774Next, let us look at Table 5 in the original paper. Here we only tabulate the values of the measures themselves, and, since the expectation is constant for each measure, the square root of the variance of the measure under randomisation — extracting values calculated above:
res <- lapply(c("MoranR", "GearyR", "Prop_unstdR", "Prop_stdR"), function(x) sapply(get(x), function(y) c(y$estimate[1], sqrt(y$estimate[3]))))
res <- t(do.call("rbind", res))
colnames(res) <- c("I", "sigma_I", "C", "sigma_C", "unstd_r", "sigma_r", "std_r", "sigma_r")
rownames(res) <- vars
print(formatC(res, format="f", digits=4), quote=FALSE)## I sigma_I C sigma_C unstd_r sigma_r std_r sigma_r
## pagval2_10 0.4074 0.1158 0.3477 0.1672 0.4559 0.1456 0.5384 0.1440
## ln_pagval2_10 0.4444 0.1183 0.3825 0.1531 0.4930 0.1490 0.5680 0.1473
## pagval10_50 0.0877 0.1143 0.5840 0.1751 0.1577 0.1436 0.1818 0.1420
## pagval50p 0.5754 0.1221 0.3925 0.1287 0.6545 0.1539 0.6795 0.1522
## cowspacre 0.5749 0.1144 0.3418 0.1745 0.5764 0.1438 0.6566 0.1421
## ln_cowspacre 0.5803 0.1186 0.4071 0.1517 0.5858 0.1493 0.6456 0.1476
## ocattlepacre 0.0244 0.1181 1.0258 0.1547 0.2775 0.1486 0.2422 0.1469
## pigspacre 0.2527 0.1159 0.6533 0.1667 0.3812 0.1458 0.4296 0.1441
## ln_pigspacre 0.2314 0.1202 0.6897 0.1417 0.3343 0.1514 0.3787 0.1497
## sheeppacre 0.0792 0.1137 0.8686 0.1778 0.2182 0.1429 0.1768 0.1412
## ln_sheeppacre 0.1117 0.1196 0.7708 0.1453 0.3125 0.1506 0.2593 0.1489
## townvillp 0.2284 0.1191 0.6148 0.1487 0.1398 0.1499 0.1987 0.1482
## carspcap 0.4914 0.1211 0.5124 0.1355 0.5394 0.1526 0.5761 0.1509
## radiopcap -0.0042 0.1188 0.8141 0.1505 0.0467 0.1495 0.0744 0.1478
## retailpcap 0.3734 0.1202 0.5267 0.1413 0.3959 0.1515 0.4066 0.1498
## psinglem30_34 0.2828 0.1207 0.6465 0.1385 0.3697 0.1520 0.3583 0.1503The values are as follows, and match the original with the exception of those for the initial version of Moran’s \(I\) in the first two columns. If we write a function implementing equations 3 and 4:
\[I = \frac{\sum_{i=1}^{n}\sum_{j=i+1}^{n}w_{ij}(x_i-\bar{x})(x_j-\bar{x})}{\sum_{i=1}^{n}(x_i - \bar{x})^2}\]
where crucially the inner summation is over \(i+1 \ldots n\), not \(1 \ldots n\), we can reproduce the values of the measure shown in the original Table 5:
oMoranf <- function(x, nb) {
z <- scale(x, scale=FALSE)
n <- length(z)
glist <- lapply(1:n, function(i) {ii <- nb[[i]]; ifelse(ii > i, 1, 0)})
lw <- nb2listw(nb, glist=glist, style="B")
wz <- lag(lw, z)
I <- (sum(z*wz)/sum(z*z))
I
}
res <- sapply(vars, function(x) oMoranf(eire_ge1[[x]], nb=lw_unstand$neighbours))
print(formatC(res, format="f", digits=4), quote=FALSE)## pagval2_10 ln_pagval2_10 pagval10_50 pagval50p cowspacre ln_cowspacre
## 0.8964 0.9776 0.1928 1.2660 1.2649 1.2766
## ocattlepacre pigspacre ln_pigspacre sheeppacre ln_sheeppacre townvillp
## 0.0536 0.5559 0.5091 0.1743 0.2458 0.5024
## carspcap radiopcap retailpcap psinglem30_34
## 1.0811 -0.0093 0.8215 0.6222The variance term given in equation 7 in the original paper is for the case of normality, not randomisation; the reference on p. 28 to equation 38 on p. 26 does not permit the reproduction of the values in the second column of Table 5. The variance equation given as equation 1.35 by Cliff and Ord (1973, 9) does not do so either, so for the time being it is not possible to say how the tabulated values were computed. Note that since the standard deviances are reproduced correctly, and can be reproduced from the second column values using the measure and its expectance, it is just a matter of establishing which formula was used, but this has so far not proved possible.
Simulating measures of spatial autocorrelation
Cliff and Ord (1969) do not conduct simulation experiments, although their sequels do, notably Cliff and Ord (1973), among many others. Simulation studies are necessarily more demanding computationally, especially if spatially autocorrelated variables are to be created, as in Cliff and Ord (1973, 146–53). In the same book, they also report the use of permutation tests, also known as Monte Carlo or Hope hypothesis testing procedures (Cliff and Ord 1973, 50–52). These procedures provided a way to examine the distribution of the statistic of interest by exchanging at random the observed values between observations, and then comparing the simulated distribution under the null hypothesis of no spatial patterning with the observed value of the statistic in question.
MoranI.boot <- function(var, i, ...) {
var <- var[i]
return(moran(x=var, ...)$I)
}
Nsim <- function(d, mle) {
n <- length(d)
rnorm(n, mle$mean, mle$sd)
}
f_bperm <- function(x, nsim, listw) {
boot(x, statistic=MoranI.boot, R=nsim, sim="permutation", listw=listw,
n=length(x), S0=Szero(listw))
}
f_bpara <- function(x, nsim, listw) {
boot(x, statistic=MoranI.boot, R=nsim, sim="parametric", ran.gen=Nsim,
mle=list(mean=mean(x), sd=sd(x)), listw=listw, n=length(x),
S0=Szero(listw))
}
nsim <- 4999
set.seed(1234)First let us define a function MoranI.boot
just to return the value of Moran’s \(I\) for variable
var and permutation index
i, and a function
Nsim to generate random samples from the
variable of interest assuming Normality. To make it easier to process
the variables in turn, we encapsulate calls to
boot in wrapper functions
f_bperm and f_bpara.
Running 4999 simulations for each of 16 for three different weights
specifications and both parametric and permutation bootstrap takes quite
a lot of time.
system.time({
MoranNb <- lapply(vars, function(x) f_bpara(x=eire_ge1[[x]], nsim=nsim, listw=nb_B))
MoranRb <- lapply(vars, function(x) f_bperm(x=eire_ge1[[x]], nsim=nsim, listw=nb_B))
Prop_unstdNb <- lapply(vars, function(x) f_bpara(x=eire_ge1[[x]], nsim=nsim, listw=lw_unstand))
Prop_unstdRb <- lapply(vars, function(x) f_bperm(x=eire_ge1[[x]], nsim=nsim, listw=lw_unstand))
Prop_stdNb <- lapply(vars, function(x) f_bpara(x=eire_ge1[[x]], nsim=nsim, listw=lw_std))
Prop_stdRb <- lapply(vars, function(x) f_bperm(x=eire_ge1[[x]], nsim=nsim, listw=lw_std))
})
res <- lapply(c("MoranNb", "MoranRb", "Prop_unstdNb", "Prop_unstdRb", "Prop_stdNb", "Prop_stdRb"), function(x) sapply(get(x), function(y) (y$t0 - mean(y$t))/sd(y$t)))
res <- t(do.call("rbind", res))
colnames(res) <- c("MoranNb", "MoranRb", "Prop_unstdNb", "Prop_unstdRb", "Prop_stdNb", "Prop_stdRb")
rownames(res) <- varsWe collate the results to compare with the analytical standard deviates under Normality and randomisation, and see that in fact the differences are not at all large, as expressed by the median absolute difference between the tables. We can also see that inferences based on a one-sided \(\alpha=0.05\) cut-off are the same for the analytical and bootstrap approaches. This indicates that we can, in general, rely on the analytical standard deviates, and that bootstrap methods will not help if assumptions underlying the measures are not met.
## pagval2_10 ln_pagval2_10 pagval10_50 pagval50p cowspacre ln_cowspacre
## 0.8964 0.9776 0.1928 1.2660 1.2649 1.2766
## ocattlepacre pigspacre ln_pigspacre sheeppacre ln_sheeppacre townvillp
## 0.0536 0.5559 0.5091 0.1743 0.2458 0.5024
## carspcap radiopcap retailpcap psinglem30_34
## 1.0811 -0.0093 0.8215 0.6222
oores <- ores - res
apply(oores, 2, mad)## MoranN MoranR Prop_unstdN Prop_unstdR Prop_stdN Prop_stdR
## 1.6470091 1.6241614 0.7681895 0.7170636 1.2615083 1.2723149## [1] FALSEThese assumptions affect the shape of the distribution of the measure
in its tails; one possibility is to use a Saddlepoint approximation to
find an equivalent to the analytical or bootstrap-based standard deviate
for inference (Tiefelsdorf 2002). The
Saddlepoint approximation requires the eigenvalues of the weights matrix
and iterative root-finding for global Moran’s \(I\), while for local Moran’s \(I_i\), analytical forms are known. Even
with this computational burden, the Saddlepoint approximation for global
Moran’s \(I\) runs quite quickly. First
we need to fit null linear models (only including an intercept) to the
variables, then apply lm.morantest.sad to the
fitted model objects:
lm_objs <- lapply(vars, function(x) lm(formula(paste(x, "~1")), data=eire_ge1))
system.time({
MoranSad <- lapply(lm_objs, function(x) lm.morantest.sad(x, listw=nb_B))
Prop_unstdSad <- lapply(lm_objs, function(x) lm.morantest.sad(x, listw=lw_unstand))
Prop_stdSad <- lapply(lm_objs, function(x) lm.morantest.sad(x, listw=lw_std))
})## user system elapsed
## 0.067 0.000 0.067
res <- sapply(c("MoranSad", "Prop_unstdSad", "Prop_stdSad"), function(x) sapply(get(x), "[[", "statistic"))
rownames(res) <- varsAlthough the analytical standard deviates (under Normality) are larger than those reached using the Saddlepoint approximation when measured by median absolute deviation, the differences do not lead to different inferences at this chosen cut-off. This reflects the fact that the shape of the distribution is very sensitive to small \(n\), but for moderate \(n\) and global Moran’s \(I\), the effects are seen only further out in the tails. The consequences for local Moran’s \(I_i\) are much stronger, because the clique of neighbours of each observation is typically very small. It is perhaps of interest that the differences are much smaller for the case of general weights than for unstandardised binary weights.
## MoranSad Prop_unstdSad Prop_stdSad
## pagval2_10 3.2903 3.1711 3.8283
## ln_pagval2_10 3.5346 3.3982 4.0441
## pagval10_50 1.0883 1.3219 1.4791
## pagval50p 4.4402 4.4258 4.9377
## cowspacre 4.4366 3.9164 4.7406
## ln_cowspacre 4.4758 3.9760 4.6493
## ocattlepacre 0.6030 2.0748 1.8642
## pigspacre 2.2611 2.7154 3.0775
## ln_pigspacre 2.1158 2.4271 2.7417
## sheeppacre 1.0250 1.7040 1.4476
## ln_sheeppacre 1.2669 2.2921 1.9731
## townvillp 2.0949 1.2079 1.5872
## carspcap 3.8500 3.6840 4.1044
## radiopcap 0.3750 0.6094 0.7906
## retailpcap 3.0663 2.8049 2.9245
## psinglem30_34 2.4649 2.6449 2.6089## MoranSad Prop_unstdSad Prop_stdSad
## 0.37142684 0.10779060 0.05650183## [1] TRUEIn addition we could choose to use the exact distribution of Moran’s \(I\), as described by Tiefelsdorf (2000); its implementation is covered in R. S. Bivand, Müller, and Reder (2009). The global case also needs the eigenvalues of the weights matrix, and the solution of a numerical integration function, but for these cases, the timings are quite acceptable.
system.time({
MoranEx <- lapply(lm_objs, function(x) lm.morantest.exact(x, listw=nb_B))
Prop_unstdEx <- lapply(lm_objs, function(x) lm.morantest.exact(x, listw=lw_unstand))
Prop_stdEx <- lapply(lm_objs, function(x) lm.morantest.exact(x, listw=lw_std))
})## user system elapsed
## 0.086 0.000 0.086
res <- sapply(c("MoranEx", "Prop_unstdEx", "Prop_stdEx"), function(x) sapply(get(x), "[[", "statistic"))
rownames(res) <- varsThe output is comparable with that of the Saddlepoint approximation, and the inferences drawn here are the same for the chosen cut-off as for the analytical standard deviates calculated under Normality.
## MoranEx Prop_unstdEx Prop_stdEx
## pagval2_10 3.2895 3.1660 3.8261
## ln_pagval2_10 3.5384 3.3979 4.0430
## pagval10_50 1.0798 1.3131 1.4745
## pagval50p 4.4568 4.4430 4.9446
## cowspacre 4.4532 3.9268 4.7453
## ln_cowspacre 4.4928 3.9875 4.6531
## ocattlepacre 0.5967 2.0611 1.8596
## pigspacre 2.2486 2.7033 3.0740
## ln_pigspacre 2.1031 2.4131 2.7380
## sheeppacre 1.0168 1.6924 1.4430
## ln_sheeppacre 1.2575 2.2779 1.9685
## townvillp 2.0822 1.1999 1.5826
## carspcap 3.8593 3.6899 4.1037
## radiopcap 0.3696 0.6054 0.7873
## retailpcap 3.0616 2.7938 2.9210
## psinglem30_34 2.4533 2.6321 2.6050## MoranEx Prop_unstdEx Prop_stdEx
## 0.37187539 0.09751723 0.05419300## [1] TRUELi, Calder, and Cressie (2007) take up the challenge in Cliff and Ord (1969, 31), to try to give the statistic a bounded fixed range. Their APLE measure is intended to approximate the spatial dependence parameter of a simultaneous autoregressive model better than Moran’s \(I\), and re-scales the measure by a function of the eigenvalues of the spatial weights matrix. APLE requires the use of row standardised weights.
vars_scaled <- lapply(vars, function(x) scale(eire_ge1[[x]], scale=FALSE))
nb_W <- nb2listw(lw_unstand$neighbours, style="W")
pre <- spatialreg:::preAple(0, listw=nb_W)
MoranAPLE <- sapply(vars_scaled, function(x) spatialreg:::inAple(x, pre))
pre <- spatialreg:::preAple(0, listw=lw_std, override_similarity_check=TRUE)
Prop_stdAPLE <- sapply(vars_scaled, function(x) spatialreg:::inAple(x, pre))
res <- cbind(MoranAPLE, Prop_stdAPLE)
colnames(res) <- c("APLE W", "APLE Gstd")
rownames(res) <- varsIn order to save time, we use the two internal functions
spatialreg:::preAple and
spatialreg:::inAple, since for each definition
of spatial weights, the same eigenvalue calculations need to be made.
The notation using the ::: operator says that
the function with named after the operator is to be found in the
namespace of the package named before the operator. The APLE values
repeat the pattern that we have already seen — for some variables, the
measured autocorrelation is very similar irrespective of spatial weights
definition, while for others, the change in the definition from binary
to general does make a difference.
## APLE W APLE Gstd
## pagval2_10 0.7702 0.6628
## ln_pagval2_10 0.7615 0.6655
## pagval10_50 0.3954 0.3446
## pagval50p 0.7519 0.7313
## cowspacre 0.8329 0.7408
## ln_cowspacre 0.8148 0.7487
## ocattlepacre 0.1468 0.4092
## pigspacre 0.6227 0.6205
## ln_pigspacre 0.5582 0.5887
## sheeppacre 0.1594 0.2841
## ln_sheeppacre 0.2046 0.4550
## townvillp 0.3442 0.2644
## carspcap 0.7140 0.6166
## radiopcap 0.0083 0.1376
## retailpcap 0.6376 0.5307
## psinglem30_34 0.4094 0.4889Odds and ends \(\ldots\)
The differences found in the case of a few variables in inference using the original binary weights, and the general weights proposed by Cliff and Ord (1969) are necessarily related to the the weights thenselves. Figures \[plot\_wts\] and \[plot\_map\] show the values of the weights in sparse matrix form, and the sums of weights by county where these sums are not identical by design. In the case of binary weights, the matrix entries are equal, but the sums up-weight counties with many neighbours.
General weights up-weight counties that are close to each other, have more neighbours, and share larger boundary proportions (an asymmetric relationship). There is a further impact of using boundary proportions, in that the boundary between the county and the exterior is subtracted, thus boosting the weights between edge counties and their neighbours, even if there are few of them. Standardised general weights up-weight further up-weight counties with few neighbours, chiefly those on the edges of the study area.
With a small data set, here with \(n=25\), it is very possible that edge and other configuration effects are relatively strong, and may impact inference in different ways. The issue of egde effects has not really been satisfactorily resolved, and should be kept in mind in analyses of data sets of this size and shape.