K nearest neighbours for spatial weights
knearneigh.RdThe function returns a matrix with the indices of points belonging to the set of the k nearest neighbours of each other. If longlat = TRUE, Great Circle distances are used. A warning will be given if identical points are found.
Arguments
- x
matrix of point coordinates, an object inheriting from SpatialPoints or an
"sf"or"sfc"object; if the"sf"or"sfc"object geometries are in geographical coordinates (sf::st_is_longlat(x) == TRUEandsf::sf_use_s2() == TRUE), s2 will be used to find the neighbours because it uses spatial indexing https://github.com/r-spatial/s2/issues/125 as opposed to the legacy method which uses brute-force- k
number of nearest neighbours to be returned; where identical points are present,
kshould be at least as large as the largest count of identical points (ifkis smaller, an error will occur whens2is used)- longlat
TRUE if point coordinates are longitude-latitude decimal degrees, in which case distances are measured in kilometers; if x is a SpatialPoints object or an
"sf"or"sfc"object, the value is taken from the object itself; longlat will overridekd_tree- use_kd_tree
logical value, if the dbscan package is available, use for finding k nearest neighbours when coordinates are planar, and when there are no identical points; from https://github.com/r-spatial/spdep/issues/38, the input data may have more than two columns if dbscan is used
Value
A list of class knn
- nn
integer matrix of region number ids
- np
number of input points
- k
input required k
- dimension
number of columns of x
- x
input coordinates
Author
Roger Bivand Roger.Bivand@nhh.no
See also
knn, dnearneigh,
knn2nb, kNN
Examples
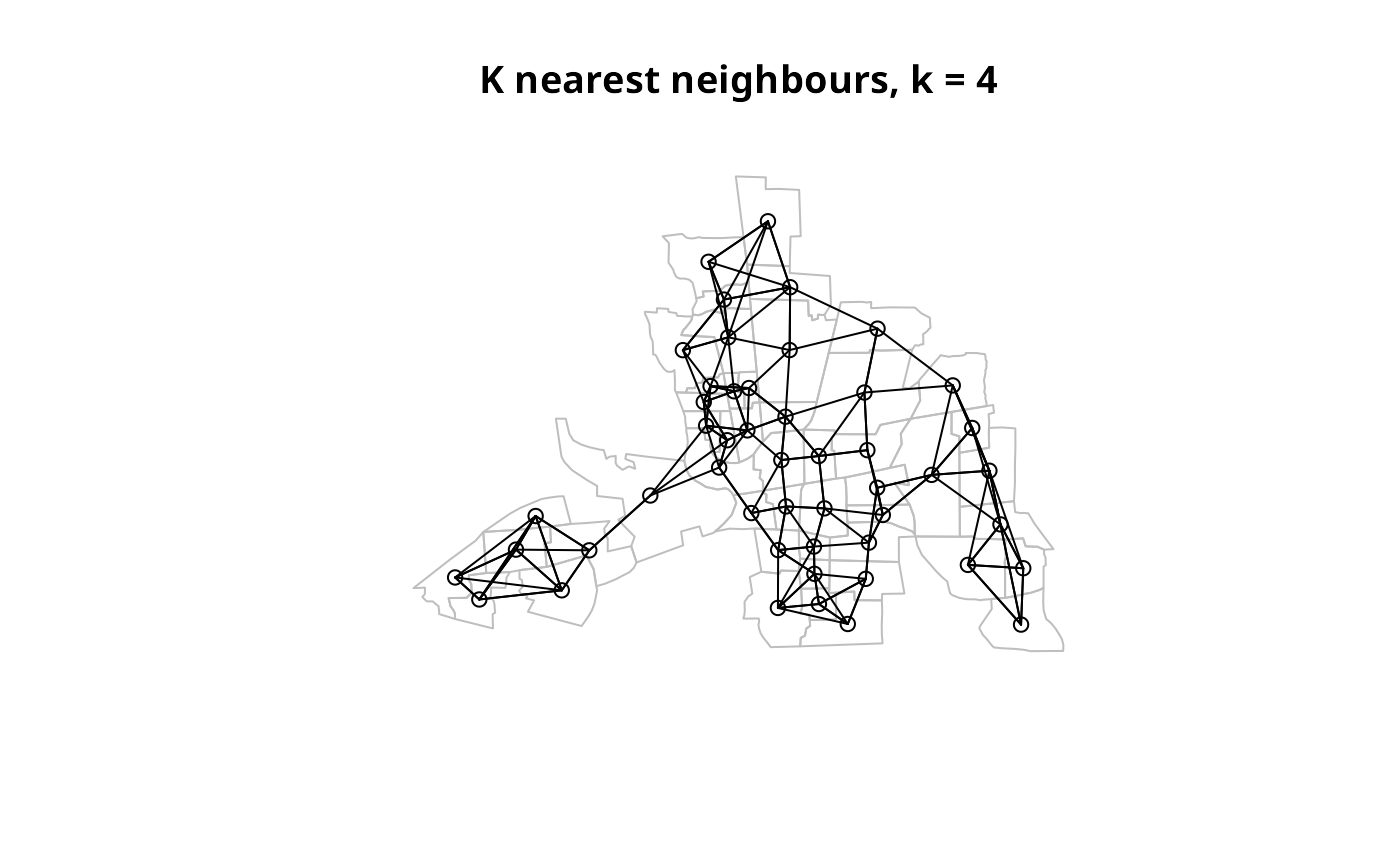
columbus <- st_read(system.file("shapes/columbus.gpkg", package="spData")[1], quiet=TRUE)
coords <- st_centroid(st_geometry(columbus), of_largest_polygon=TRUE)
col.knn <- knearneigh(coords, k=4)
plot(st_geometry(columbus), border="grey")
plot(knn2nb(col.knn), coords, add=TRUE)
title(main="K nearest neighbours, k = 4")
 data(state)
us48.fipsno <- read.geoda(system.file("etc/weights/us48.txt",
package="spdep")[1])
if (as.numeric(paste(version$major, version$minor, sep="")) < 19) {
m50.48 <- match(us48.fipsno$"State.name", state.name)
} else {
m50.48 <- match(us48.fipsno$"State_name", state.name)
}
xy <- as.matrix(as.data.frame(state.center))[m50.48,]
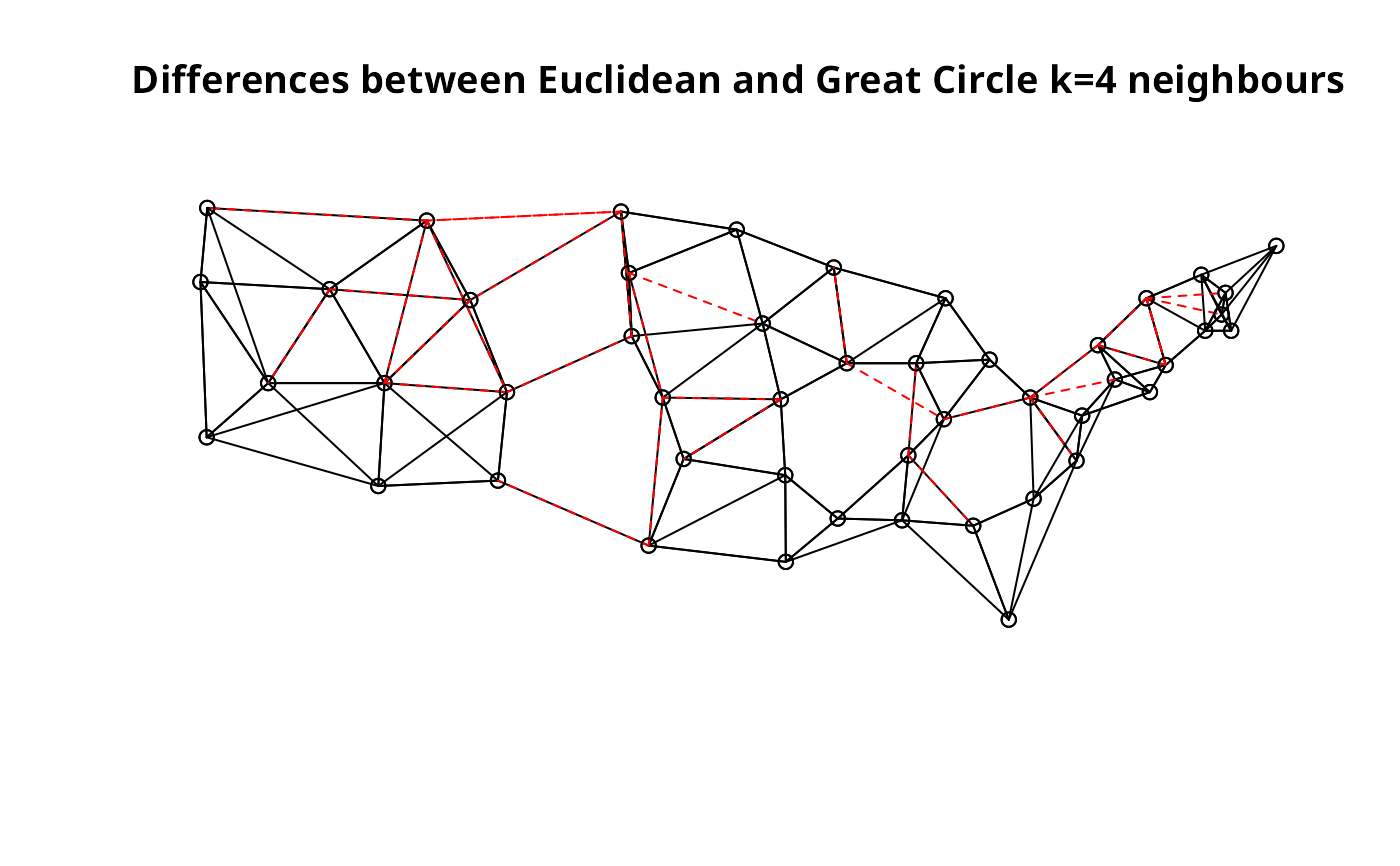
llk4.nb <- knn2nb(knearneigh(xy, k=4, longlat=FALSE))
gck4.nb <- knn2nb(knearneigh(xy, k=4, longlat=TRUE))
plot(llk4.nb, xy)
plot(diffnb(llk4.nb, gck4.nb), xy, add=TRUE, col="red", lty=2)
#> Warning: neighbour object has 22 sub-graphs
title(main="Differences between Euclidean and Great Circle k=4 neighbours")
data(state)
us48.fipsno <- read.geoda(system.file("etc/weights/us48.txt",
package="spdep")[1])
if (as.numeric(paste(version$major, version$minor, sep="")) < 19) {
m50.48 <- match(us48.fipsno$"State.name", state.name)
} else {
m50.48 <- match(us48.fipsno$"State_name", state.name)
}
xy <- as.matrix(as.data.frame(state.center))[m50.48,]
llk4.nb <- knn2nb(knearneigh(xy, k=4, longlat=FALSE))
gck4.nb <- knn2nb(knearneigh(xy, k=4, longlat=TRUE))
plot(llk4.nb, xy)
plot(diffnb(llk4.nb, gck4.nb), xy, add=TRUE, col="red", lty=2)
#> Warning: neighbour object has 22 sub-graphs
title(main="Differences between Euclidean and Great Circle k=4 neighbours")
 summary(llk4.nb, xy, longlat=TRUE, scale=0.5)
#> Neighbour list object:
#> Number of regions: 48
#> Number of nonzero links: 192
#> Percentage nonzero weights: 8.333333
#> Average number of links: 4
#> Non-symmetric neighbours list
#> Link number distribution:
#>
#> 4
#> 48
#> 48 least connected regions:
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 with 4 links
#> 48 most connected regions:
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 with 4 links
summary(gck4.nb, xy, longlat=TRUE, scale=0.5)
#> Neighbour list object:
#> Number of regions: 48
#> Number of nonzero links: 192
#> Percentage nonzero weights: 8.333333
#> Average number of links: 4
#> Non-symmetric neighbours list
#> Link number distribution:
#>
#> 4
#> 48
#> 48 least connected regions:
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 with 4 links
#> 48 most connected regions:
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 with 4 links
#xy1 <- SpatialPoints((as.data.frame(state.center))[m50.48,],
# proj4string=CRS("+proj=longlat +ellps=GRS80"))
#gck4a.nb <- knn2nb(knearneigh(xy1, k=4))
#summary(gck4a.nb, xy1, scale=0.5)
xy1 <- st_as_sf((as.data.frame(state.center))[m50.48,], coords=1:2,
crs=st_crs("OGC:CRS84"))
old_use_s2 <- sf_use_s2()
sf_use_s2(TRUE)
system.time(gck4a.nb <- knn2nb(knearneigh(xy1, k=4)))
#> user system elapsed
#> 0.011 0.000 0.011
summary(gck4a.nb, xy1, scale=0.5)
#> Neighbour list object:
#> Number of regions: 48
#> Number of nonzero links: 192
#> Percentage nonzero weights: 8.333333
#> Average number of links: 4
#> Non-symmetric neighbours list
#> Link number distribution:
#>
#> 4
#> 48
#> 48 least connected regions:
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 with 4 links
#> 48 most connected regions:
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 with 4 links
sf_use_s2(FALSE)
#> Spherical geometry (s2) switched off
system.time(gck4a.nb <- knn2nb(knearneigh(xy1, k=4)))
#> user system elapsed
#> 0.007 0.000 0.007
summary(gck4a.nb, xy1, scale=0.5)
#> Neighbour list object:
#> Number of regions: 48
#> Number of nonzero links: 192
#> Percentage nonzero weights: 8.333333
#> Average number of links: 4
#> Non-symmetric neighbours list
#> Link number distribution:
#>
#> 4
#> 48
#> 48 least connected regions:
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 with 4 links
#> 48 most connected regions:
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 with 4 links
sf_use_s2(old_use_s2)
#> Spherical geometry (s2) switched on
# https://github.com/r-spatial/spdep/issues/38
run <- FALSE
if (require("dbscan", quietly=TRUE)) run <- TRUE
if (run) {
set.seed(1)
x <- cbind(runif(50), runif(50), runif(50))
out <- knearneigh(x, k=5)
knn2nb(out)
}
#> Neighbour list object:
#> Number of regions: 50
#> Number of nonzero links: 250
#> Percentage nonzero weights: 10
#> Average number of links: 5
#> Non-symmetric neighbours list
if (run) {
# fails because dbscan works with > 2 dimensions but does
# not work with duplicate points
try(out <- knearneigh(rbind(x, x[1:10,]), k=5))
}
#> Warning: knearneigh: identical points found
#> Warning: knearneigh: kd_tree not available for identical points
#> Error in knearneigh(rbind(x, x[1:10, ]), k = 5) :
#> kd_tree required for more than 2 dimensions
summary(llk4.nb, xy, longlat=TRUE, scale=0.5)
#> Neighbour list object:
#> Number of regions: 48
#> Number of nonzero links: 192
#> Percentage nonzero weights: 8.333333
#> Average number of links: 4
#> Non-symmetric neighbours list
#> Link number distribution:
#>
#> 4
#> 48
#> 48 least connected regions:
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 with 4 links
#> 48 most connected regions:
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 with 4 links
summary(gck4.nb, xy, longlat=TRUE, scale=0.5)
#> Neighbour list object:
#> Number of regions: 48
#> Number of nonzero links: 192
#> Percentage nonzero weights: 8.333333
#> Average number of links: 4
#> Non-symmetric neighbours list
#> Link number distribution:
#>
#> 4
#> 48
#> 48 least connected regions:
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 with 4 links
#> 48 most connected regions:
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 with 4 links
#xy1 <- SpatialPoints((as.data.frame(state.center))[m50.48,],
# proj4string=CRS("+proj=longlat +ellps=GRS80"))
#gck4a.nb <- knn2nb(knearneigh(xy1, k=4))
#summary(gck4a.nb, xy1, scale=0.5)
xy1 <- st_as_sf((as.data.frame(state.center))[m50.48,], coords=1:2,
crs=st_crs("OGC:CRS84"))
old_use_s2 <- sf_use_s2()
sf_use_s2(TRUE)
system.time(gck4a.nb <- knn2nb(knearneigh(xy1, k=4)))
#> user system elapsed
#> 0.011 0.000 0.011
summary(gck4a.nb, xy1, scale=0.5)
#> Neighbour list object:
#> Number of regions: 48
#> Number of nonzero links: 192
#> Percentage nonzero weights: 8.333333
#> Average number of links: 4
#> Non-symmetric neighbours list
#> Link number distribution:
#>
#> 4
#> 48
#> 48 least connected regions:
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 with 4 links
#> 48 most connected regions:
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 with 4 links
sf_use_s2(FALSE)
#> Spherical geometry (s2) switched off
system.time(gck4a.nb <- knn2nb(knearneigh(xy1, k=4)))
#> user system elapsed
#> 0.007 0.000 0.007
summary(gck4a.nb, xy1, scale=0.5)
#> Neighbour list object:
#> Number of regions: 48
#> Number of nonzero links: 192
#> Percentage nonzero weights: 8.333333
#> Average number of links: 4
#> Non-symmetric neighbours list
#> Link number distribution:
#>
#> 4
#> 48
#> 48 least connected regions:
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 with 4 links
#> 48 most connected regions:
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 with 4 links
sf_use_s2(old_use_s2)
#> Spherical geometry (s2) switched on
# https://github.com/r-spatial/spdep/issues/38
run <- FALSE
if (require("dbscan", quietly=TRUE)) run <- TRUE
if (run) {
set.seed(1)
x <- cbind(runif(50), runif(50), runif(50))
out <- knearneigh(x, k=5)
knn2nb(out)
}
#> Neighbour list object:
#> Number of regions: 50
#> Number of nonzero links: 250
#> Percentage nonzero weights: 10
#> Average number of links: 5
#> Non-symmetric neighbours list
if (run) {
# fails because dbscan works with > 2 dimensions but does
# not work with duplicate points
try(out <- knearneigh(rbind(x, x[1:10,]), k=5))
}
#> Warning: knearneigh: identical points found
#> Warning: knearneigh: kd_tree not available for identical points
#> Error in knearneigh(rbind(x, x[1:10, ]), k = 5) :
#> kd_tree required for more than 2 dimensions